bayesGO is a Bayesian hierarchical model that simultaneously identifies pathway-modulating genes based on the literature mining data and facilitates interpreting functions of these new genes using Gene Ontology terms. This approach allows rigorous inference of gene-gene relationships based on the literature mining data while the GAIL web interface allows users to implement dynamic and interactive exploratory analyses.
Download
The R package bayesGO implements this statistical model and provides simple and user-friendly interface for its statistical inference. The 'P-Value Matrix' output from the GAIL web interface can be used as input for this software. Note, bayesGO requires JAGS. You can download JAGS and bayesGO through following links:
Usage
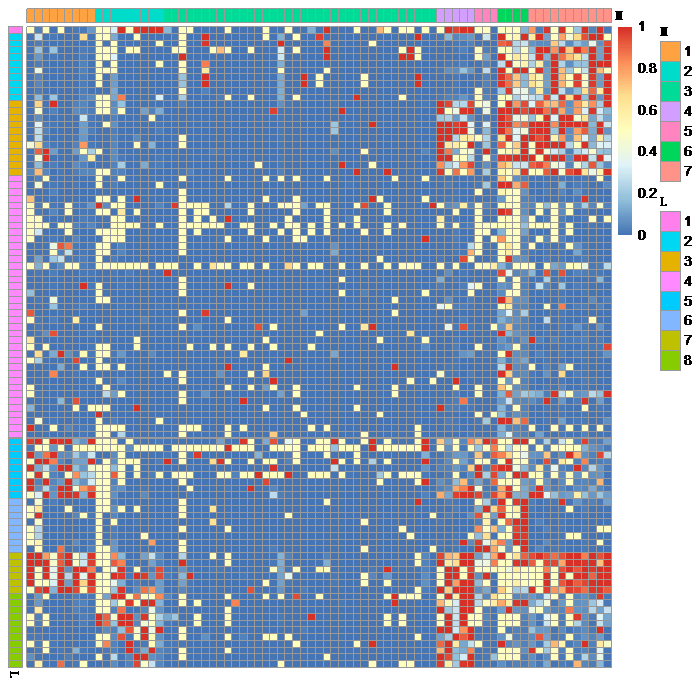
After you download the ‘P-Value Matrix’ output from the GAIL web interface, you can load this file into the R environment using the R function read.csv() and the R function bayesGO() fits the bayesGO model by taking this data as input. Then, the R function predict() implements clustering of genes and GO terms and identifies association between genes and GO terms. Finally, the R function plot() visualizes the analysis results as below, where column and row side bars show gene and GO term cluster indices and red colors within the heatmap indicates stronger association between genes and GO terms. Please check Yu et al. (2018) for the step-by-step analysis guideline and Chung et al. (2017) for more details about the statistical model.